
WHAT IS SBGN
SPECIFICATIONS
SOFTWARE
EVENTS
FAQ
HOW TO CITE
ABOUT
CONTACT
Learning SBGN
Examples
Reference Cards
Published Maps
Symbol Highlights
Figure to SBGN
Competition
Development Wiki
Learning SBGN
This page offers a brief introduction to the SBGN, and, in particular, to the Process Description language as the currently most used language of the SBGN standard. Redrawing a small example diagram will help with a quick start, and examples of the SBGN Bricks will help to align biological concepts with their graphical expressions.
The three languages of SBGN
There are three complementary languages in SBGN: Activity Flow (AF), Process Description (PD) and Entity Relationship (ER).
In the figure, the same biological system is shown in different representations depending on the concepts used to describe this system (Le Novère, 2015, doi:10.1038/nrg3885). Note the same set of four proteins in all cases. In the Process Description, ELK1 is shown in three different states, with ELK1 sumoylation (SUMO) and phosphorylation (P) processes.
Next, to avoid any confusion, we concentrate only on the Process Description (PD) language.
Process in SBGN PD
The process glyph is the key element for understanding of the SBGN PD language:
Reading a PD diagram is much simpler if it is seen as a collection of interconnected processes. Represented in PD, a biological process includes 1) incoming consumption link(s) to the process, 2) production link(s) from the process, and often 3) regulatory link(s) to the process, for example stimulation or inhibition.
 SBGN-ML Newt |
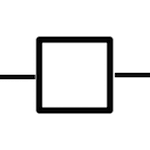
The above example shows the phosphorylation of glucose by hexokinase. The process consumes glucose and ATP to produce glucose-6P and ADP. Glucose and ATP are linked to the process by consumption arcs; glucose-6P and ADP by production arcs; and the hexokinase by a catalysis arc.
SBGN PD Bricks
The SBGN Bricks project presents a tamplate-based approach that makes it simpler to learn and start applying the standard without necesserily being familiar with all the specifications. Here are some illustrations of how biological concepts such as metabolic reaction or complex formation can be reflected in the SBGN Process Description language (Junker et al., 2012, doi:10.1016/j.tibtech.2012.08.003).
The example bricks are developed using the SBGN-ED add-on of VANTED editor (desktop application). All the bricks are available for downloading in SBGN-ML format and can be opened online in Newt (online editor). The Krayon editor (desktop application) supports the SBGN Bricks and the available templates can be used to design a diagram by adding one brick after another. To download Krayon please use the Releases page.
Metabolic processes
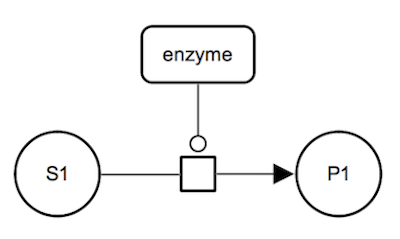 SBGN-ML Newt |
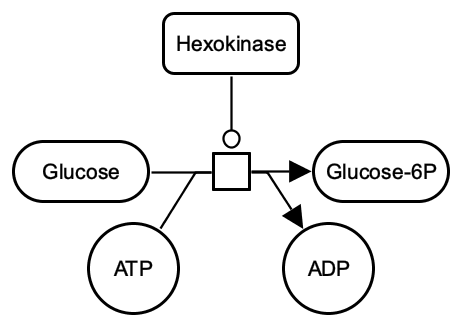
Catalysis: irreversible reacton. The enzyme catalyses an irreversible metabolic process which consumes substrate S1 and produces product P1. The enzyme is a represented as a macromolecule connected to the process glyph by a catalysis arc. The substrate and the product of the biochemical reaction are represented by simple chemical glyphs. |
 SBGN-ML Newt |
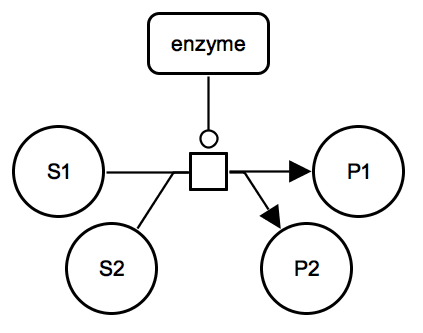
Catalysis: multiple substrates and products. The enzyme catalyses an irreversible metabolic process which consumes two substrates S1 and S2 and produces two products P1 and P2. |
 SBGN-ML Newt |
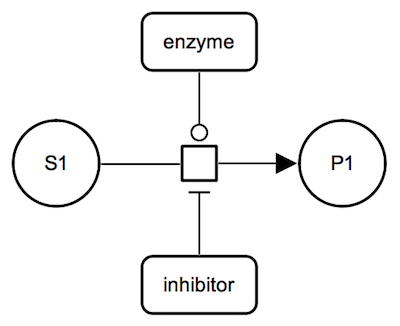
Inhibition: irreversible metabolic reaction. The inhibitor, a proteins shown with a macromolecule glyph, is connected to the process glyph by an inhibition arc. |
Signalling processes
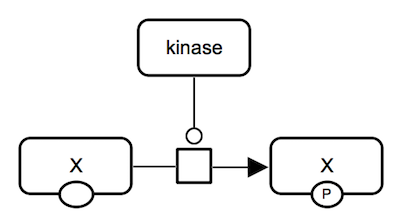 SBGN-ML Newt |
Protein phosphorylation. A kinase protein catalyzes an irreversible reaction which consumes unphosphorylated protein X and produces phosphorylated protein X. All proteins involved are represented by macromolecules. |
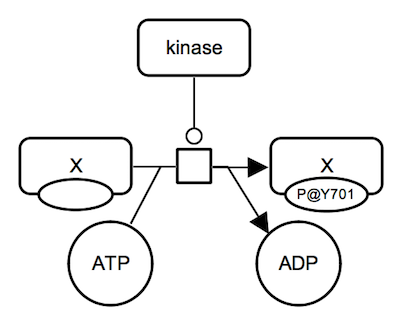 SBGN-ML Newt |
Protein phosphorylation (with more details captured). A kinase protein catalyzes an irreversible reaction which consumes unphosphorylated protein X and ATP and produces phosphorylated protein X and ADP. All proteins involved are represented by macromolecule glyphs. State variable auxiliary glyphs are used to indicate the phosphorylation state: "P@Y701" means "phosphorylated at tyrosine 701" (one-letter amino acid code). Instead of empty state, "@Y701" without "P" can be used to indicate the position. ATP and ADP are represented as simple chemicals. |
An extended collection of the PD patterns is available at the SBGN Bricks website.
iNOS pathway example
After simply redrawing the following example you will be much more familiar with how the SBGN PD language works. This was proved to be very useful in various tutorials and university courses. The example pathway diagram is comparatively small but includes many types of biological processes and the corresponding elements of the graphical notation.
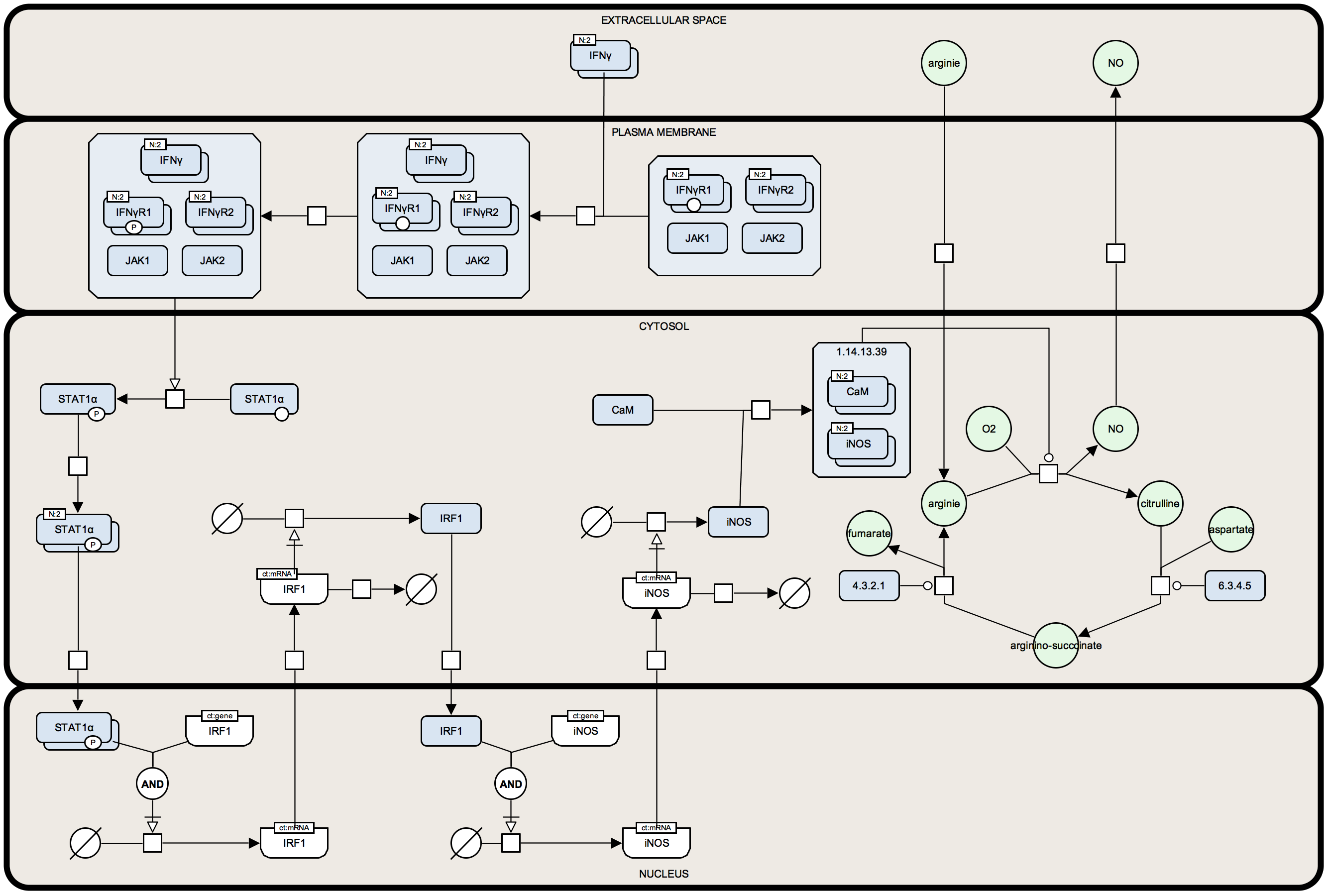
The diagram can be redrawn in any application that supports the required shapes, for example, in PowerPoint, InkScape or Adobe Illustrator. In GraphML format the diagram can be developed in yEd Graph Editor (desktop application) that since version 3.17.1 provides a palette section for SBGN.
To build the diagram and generate an SBGN-ML file, a dedicated software should be used, for example Newt (online editor) or SBGN-ED add-on for VANTED (desktop application). The SBGN Validation Tool can be used to verify the quality of your diagram in VANTED (Tools tab of the SBGN-ED add-on).
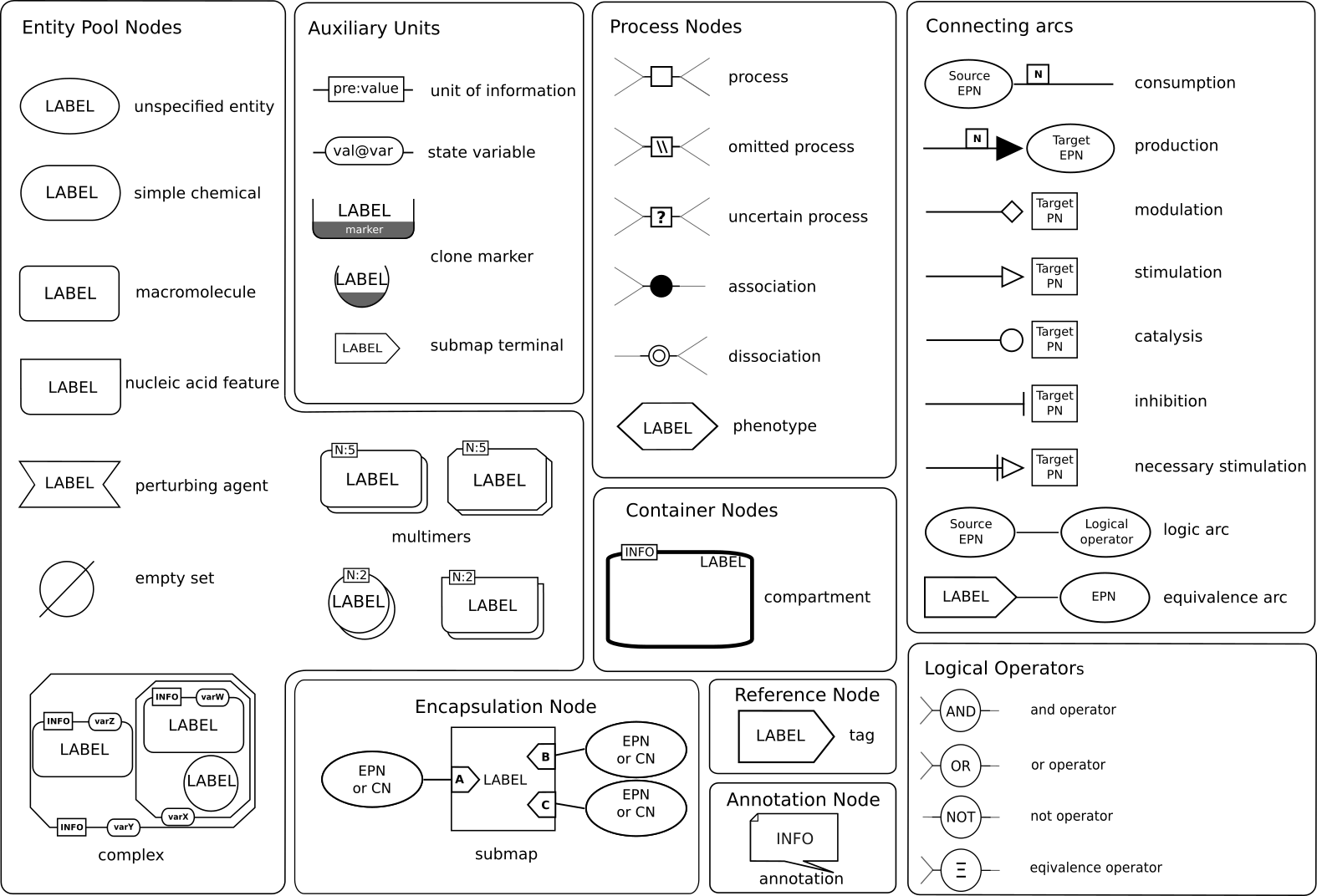
SBGN PD Reference Card
Reading list
- Le Novère et al. The Systems Biology Graphical Notation. Nat Biotechnol. 2009. doi:10.1038/nbt.1558
- Touré et al. Quick tips for creating effective and impactful biological pathways using the Systems Biology Graphical Notation. PLoS Computat Biol. 2018. doi:10.1371/journal.pcbi.1005740
- Le Novère. Quantitative and logic modelling of molecular and gene networks. Nat Rev Genet. 2015. doi:10.1038/nrg3885
Tutorials
Tutorial materials can be found at our Education Resource repository.
Examples
The Examples page provides a number of SBGN diagrams in three SBGN languages.
Reference cards
The Reference Cards page offers reference cards in various formats.
Additional materials
For more advanced questions please use FAQ, in particular FAQ Process Description.
For questions and comments please contact the SBGN editors at
sbgn-editors@googlegroups.com