
WHAT IS SBGN
SPECIFICATIONS
SOFTWARE
EVENTS
FAQ
HOW TO CITE
ABOUT
CONTACT
Learning SBGN
Examples
Reference Cards
Published Maps
Symbol Highlights
Figure to SBGN
Competition
Development Wiki
Symbols
- PD Stimulation
- PD Inhibition
- PD Process
- PD Consumption Production Reversible
- PD Macromolecule
- PD Simple Chemical
- PD Catalysis
- PD Nucleic Acid Feature
- PD Complex
- AF and PD Compartment
- PD Clone Marker
- AF Biological Activity
- AF Unit Of Information for BA
- PD Perturbing Agent
- PD Unspecified Entity
- PD Source and Sink
- PD ER and AF And
- PD ER and AF Or
- PD ER and AF Not
PD Stimulation
By Nicolas Le Novère (Nov 2012)
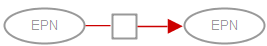
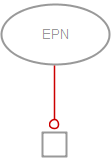
A stimulation affects positively the flux of a process (its speed). This stimulation can be, for instance, a catalysis or a positive allosteric regulation. In SBGN Process Description language, the stimulation of a process is represented by the stimulation arc (Note that catalysis exists as an independent symbol in SBGN PD). The target extremity of a stimulation carries an empty arrowhead (note that the stimulation arcs of all three SBGN languages have the same appearance). A stimulation arc starts from an entity pool node (labelled “EPN” in the figure below, representing for instance a pool of molecules) and ends on a process node (labelled “PN” in the figure below, representing for instance a reaction). In the drawing below, the stimulation arc is coloured in red:
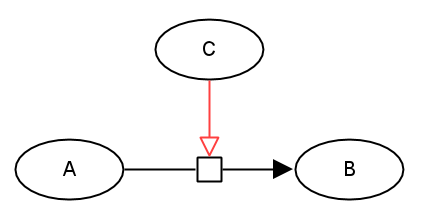
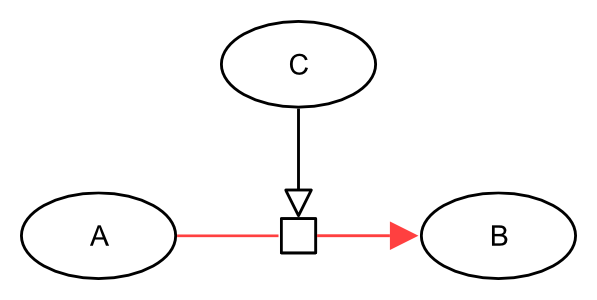
The following generic example describes an entity C that stimulates the process transforming an entity A into an entity B.
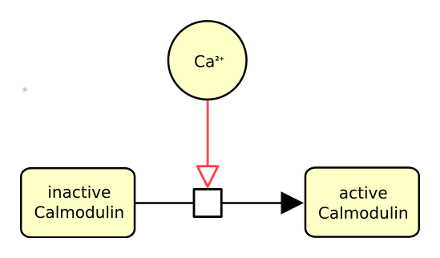
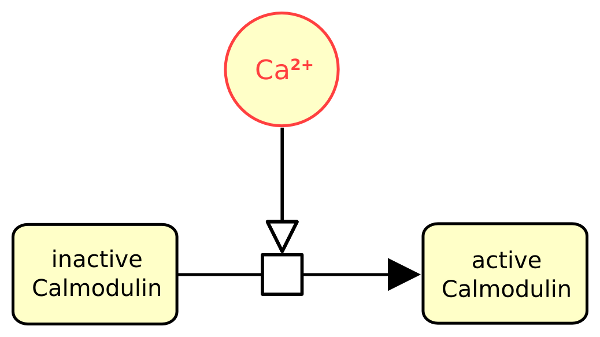
The following actual example describes calcium stimulating the conversion of inactive calmodulin into active one.
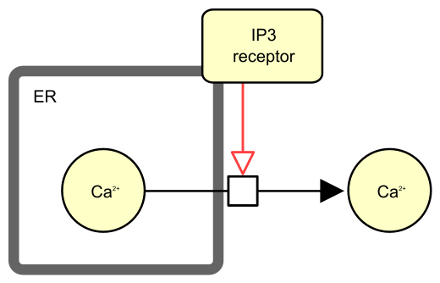
The following actual example describes the stimulation of calcium excretion from the endoplasmic reticulum by the inositol 3 phosphate receptor.
Download the stimulation arc image as (from the more semantic to the more generic): [ SBGN-ML ] [ GML ] [ GraphML ] [ SVG ] [ Open Document Format ] [ PowerPoint ] [ PDF ] [ PNG ]
PD Inhibition
By Nicolas Le Novère (Dec 2012)
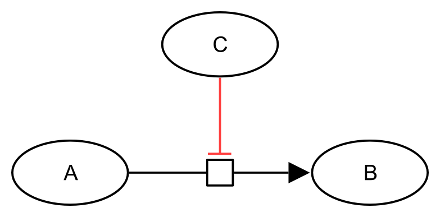
An inhibition affects negatively the flux of a process (its speed). This inhibition can be, for instance, the effect of a channel blocker or a negative allosteric regulation. In SBGN Process Description language, the inhibition of a process is represented by the inhibition arc (note that the inhibition arcs of all three SBGN languages have the same appearance). The target extremity of an inhibition carries a bar perpendicular to the end of the arc. An inhibition arc starts from an entity pool node (labelled “EPN” in the figure below, representing for instance a pool of molecules) and ends on a process node (labelled “PN” in the figure below, representing for instance a reaction). In the drawing below, the inhibition arc is coloured in red:

The following generic example describes an entity C that inhibits the process transforming an entity A into an entity B.
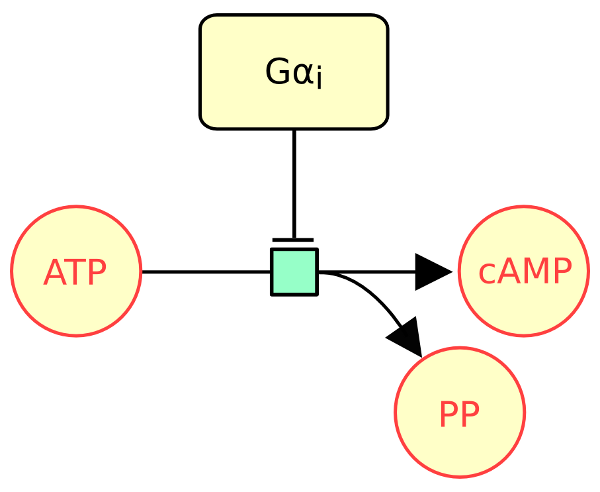
The following actual example describes the protein Gαi inhibiting the conversion of ATP into cAMP.
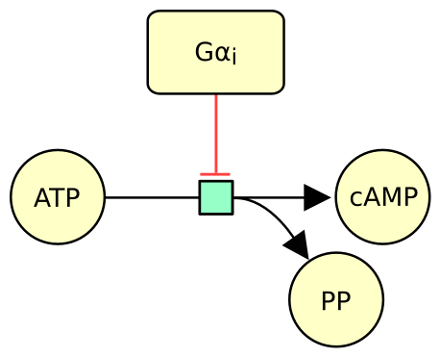
Download the inhibition arc image as (from the more semantic to the more generic): [ SBGN-ML ] [ GML ] [ GraphML ] [ SVG ] [ Open Document Format ] [ PowerPoint ] [ PDF ] [ PNG ]
PD Process
By Nicolas Le Novère (Jan 2013)
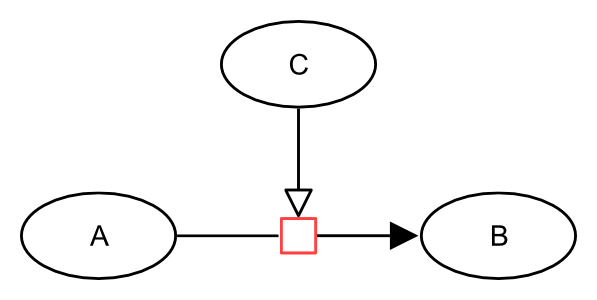
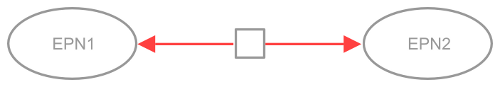
A process is something that happens, involving one or more physical entities, and modifies the structure, location or free energy of at least one of the participants. Examples would be biochemical reactions or transports. In SBGN Process Description language, a process is represented by the process node, an unlabelled square. A process node is connected to the entity participating (entity pool node, labelled “EPN” in the figure below), via arcs representing consumption, production or modulation. In the drawing below, the process node is coloured in red:

The following generic example describes an entity C that stimulates the process transforming an entity A into an entity B.
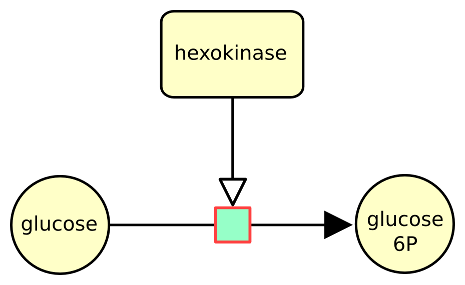
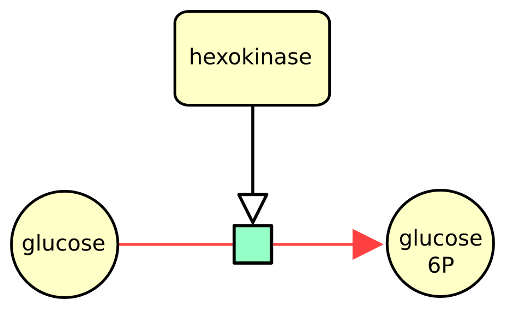
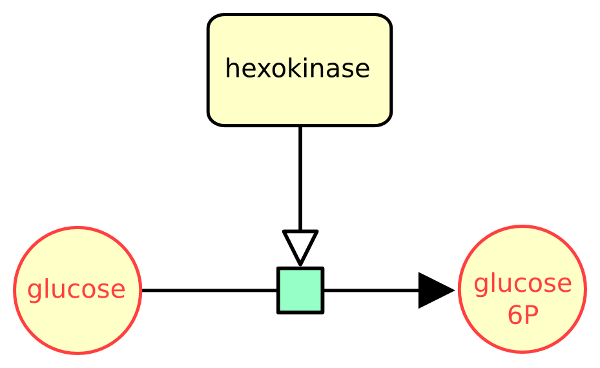
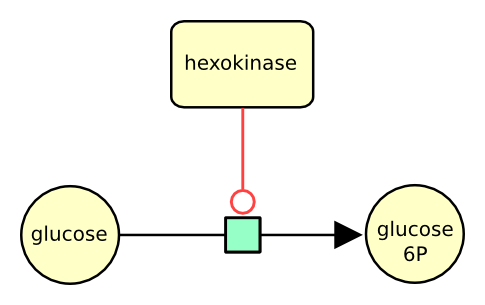
The following example describes the conversion of glucose into glucose 6 phosphate, stimulated by the enzyme hexokinase.
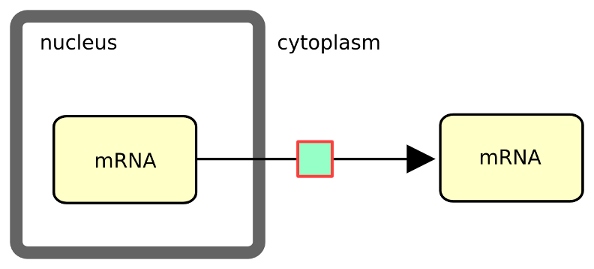
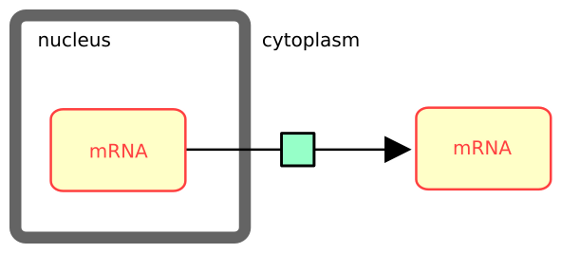
The following example describes the transport of messenger RNA from the nucleus to the cytoplasm. Note that in SBGN, the containing compartment is part of the identity of an EPN. Therefore one does not need to differentiate the two mRNA pools via their labels (e.g. mRNA_nuc and mRNA_cyt).
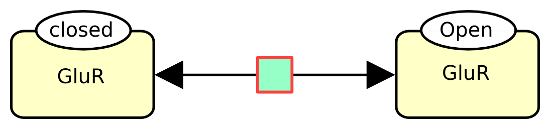
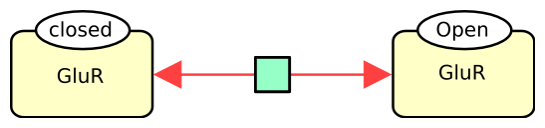
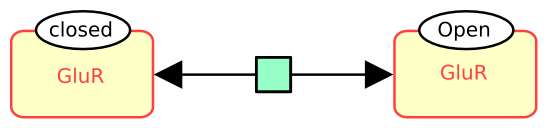
The following example describes the reversible conversion of the glutamate ionotropic receptor between closed and open states.
Download the process node image as (from the more semantic to the more generic): [ SBGN-ML ] [ GML ] [ GraphML ] [ SVG ] [ Open Document Format ] [ PowerPoint ] [ PDF ] [ PNG ]
PD Consumption Production Reversible
By Nicolas Le Novère (Feb 2013)
A process transforms one or several physical entities into one or several others. Examples would be biochemical reactions converting chemicals into others or transport reactions moving chemicals from one compartment to another. In SBGN Process Description language, the use of reactants is represented by the consumption arc, a simple arc without specific ends. The generation of products is represented by the production arc, an arc ending by a filled arrowhead. In the drawing below, the consumption and production arcs are coloured in red:
The following generic example describes an entity C that stimulates the process consuming an entity A and producing an entity B.
A reaction or a transport can be reversible. In that case, both side of the process are linked to production arcs:
The following example describes the consumption of glucose to produce glucose 6 phosphate, stimulated by the enzyme hexokinase.
The following example describes the reversible conversion of the glutamate ionotropic receptor between closed and open states.
Download the consumption and production arcs images as (from the more semantic to the more generic): [ SBGN-ML ] [ GML ] [ GraphML ] [ SVG ] [ Open Document Format ] [ PowerPoint ] [ PDF ] [ PNG ]
PD Macromolecule
By Nicolas Le Novère (Mar 2013)
Many biological processes involve macromolecules: biochemical substances that are built up from the covalent linking of pseudo-identical units. Examples of macromolecules include proteins, nucleic acids (RNA, DNA), and polysaccharides (glycogen, cellulose, starch, etc.). Attempting to define a separate glyph for all of these different molecules would lead to an explosion of symbols in SBGN, so instead, SBGN Process Description Level 1 defines only one glyph for all macromolecules. The same glyph is to be used for a protein, a nucleic acid, a complex sugar, and so on. The exact nature of a particular macromolecule in a map is then clarified using its label and decorations. A macromolecule is represented by a rectangular container with rounded corners.

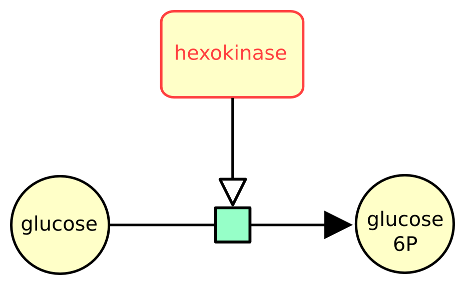
The following example describes the conversion of glucose into glucose 6 phosphate, stimulated by the enzyme hexokinase, represented as a macromolecule.
The following example describes the transport of messenger RNA from the nucleus to the cytoplasm. One does not need to differentiate the two mRNA pools via their labels (e.g. mRNA_nuc and mRNA_cyt) since in SBGN, the containing compartment is part of the identity of a pool of entities.
Macromolecules can be decorated by state variables or unit of information, as we will present in future symbols of the month. For now, let’s just say that the state of a macromolecule (that is the values of all its state variables) is part of its identity. The following example describes the reversible conversion of the glutamate ionotropic receptor between closed and open states.
Download the macromolecule node image as (from the more semantic to the more generic): [ SBGN-ML ] [ GML ] [ GraphML ] [ SVG ] [ Open Document Format ] [ PowerPoint ] [ PDF ] [ PNG ]
PD Simple Chemical
By Nicolas Le Novère (Apr 2013)
In SBGN Process Descriptions, a simple chemical is defined as the opposite of a macromolecule: it is a chemical compound that is not formed by the covalent linking of pseudo-identical residues. Examples of simple chemicals are an atom, a monoatomic ion, a salt, a radical, a solid metal, a crystal, etc. A simple chemical is represented by a circular container.

The following example describes the conversion of glucose into glucose 6 phosphate, stimulated by the enzyme hexokinase, represented as a macromolecule.
The following example describes calcium stimulating the conversion of inactive calmodulin into active one.
The following example describes the protein Gαi inhibiting the conversion of ATP into cAMP.
Download the macromolecule node image as (from the more semantic to the more generic): [ SBGN-ML ] [ GML ] [ GraphML ] [ SVG ] [ Open Document Format ] [ PowerPoint ] [ PDF ] [ PNG ]
PD Catalysis
By Nicolas Le Novère (Jun 2013)
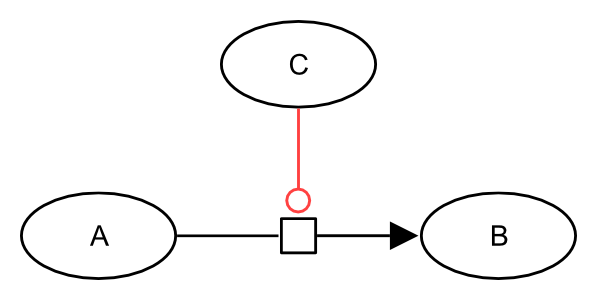
A catalysis is a particular case of stimulation, where the effector affects positively the flux of a process represented by the target process. The positive effect on the process is due to the lowering of the activation energy of a reaction. The target extremity of a catalysis carries an empty circle.
In SBGN Process Description language, the catalysis of a process is represented by the catalysis arc. The target extremity of a catalysis carries an empty circle. A catalysis arc starts from an entity pool node (labelled “EPN” in the figure below, representing for instance a pool of molecules) and ends on a process node (labelled “PN” in the figure below, representing for instance a reaction). In the drawing below, the catalysis arc is coloured in red:
The following generic example describes an entity C that catalyses the process transforming an entity A into an entity B.
The following actual example describes the conversion of glucose into glucose 6 phosphate, catalysed by the enzyme hexokinase.
Download the process node image as (from the more semantic to the more generic): [ SBGN-ML ] [ GML ] [ GraphML ] [ SVG ] [ Open Document Format ] [ PowerPoint ] [ PDF ] [ PNG ]
PD Nucleic Acid Feature
By Nicolas Le Novère (Oct 2013)
The Nucleic Acid Feature construct in SBGN is meant to represent a fragment of a macromolecule carrying genetic information. A common use for this construct is to represent a gene or a transcript. The macromolecule would be the entire nucleic acid molecule, such as an entire chromatide. The Nucleic acid feature has two round corners, to remind it is carried by a macromolecule, and two right angle corners, to remind it is essentially a unit of information rather than a bona fide molecule.

The following example describes the synthesis of a protein from amino-acids, triggered (“necessary stimulated”) by mRNA.
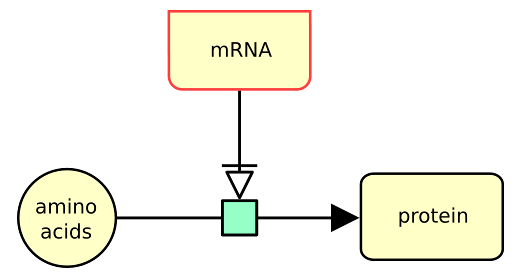
Download the nucleic acid feature node image as (from the more semantic to the more generic): [ SBGN-ML ] [ GML ] [ GraphML ] [ SVG ] [ Open Document Format ] [ PowerPoint ] [ PDF ] [ PNG ]
PD Complex
By Nicolas Le Novère (Jan 2014)
A complex node represents a biochemical entity composed of other biochemical entities, whether macromolecules, simple chemicals, multimers, or other complexes, linked by non-covalent bounds. The complex construct in SBGN possesses its own symbol, which is a rectangle with cut-corners (an octagonal box with sides of two different lengths). This symbol can be decorated with the symbols of its components. In this case, the label of the complex is optional.
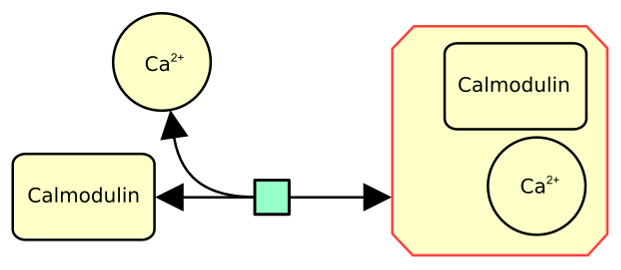
The following example describes the reversible binding of calcium (represented as a simple chemical) on calmodulin (represented as a macromolecule).
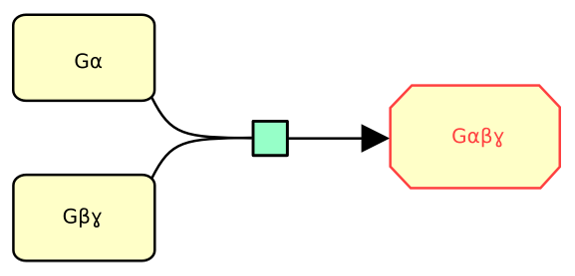
The following example describes the binding the subunit alpha with subunits beta and gamma of the heterotrimeric G-protein.
Download the complex node image as (from the more semantic to the more generic): [ SBGN-ML ] [ GML ] [ GraphML ] [ SVG ] [ Open Document Format ] [ PowerPoint ] [ PDF ] [ PNG ]
AF and PD Compartment
by Vasundra Touré (Jan 2016)
A compartment is a container node found in SBGN PD and SBGN AF. It is a logical or physical structure that contains entity pool nodes (EPNs). One EPN belongs to one compartment. Thus, two “same” entities located in two different compartments are actually two different pools. The compartment has its own symbol: it is an enclosed structure with thick borders that can have any geometry. It should carry a label to characterise the compartment’s name.

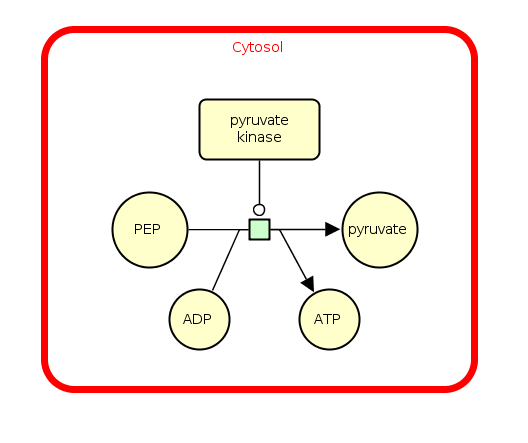
The following example represents the consumption of phosphoenolpyruvate (PEP) to produce pyruvate in the cytosol compartment. The reaction is stimulated by the enzyme pyruvate kinase and produces a molecule of energy ATP.
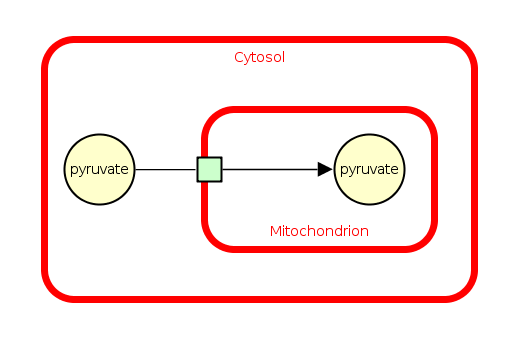
Compartments can overlap but that does not always imply that the overlapped compartment contains the overlapping one. Also, the overlapped part of a compartment should not contain any object which could be covered by an overlapping compartment. The following example shows the transport of pyruvate from the cytosol to the mitochondrion. Here, the cytosol is overlapped by the mitochondrion / the mitochondrion is overlapping the cytosol.
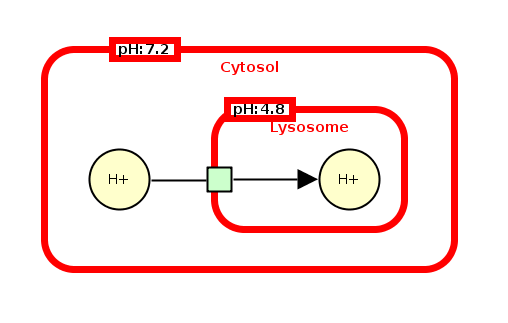
Finally, compartments can hold units of information that will for instance give additional information about the physical environment of the compartment (pH, temperature, voltage, etc.). The following example represents the lysosome compartment with a pH of 4,8 and the cytosol compartment with a pH of 7,2. These units of information show that the lysosome is more acidic than the cytosol.
Download the compartment node image as (from the more semantic to the more generic):
[ SBGN-ML ] [ GML ] [ GraphML ] [ SVG ] [ Open Document Format ] [ PowerPoint ] [ PDF ] [ PNG ]
PD Clone Marker
by Vasundra Touré (Feb 2016)
A clone marker is an auxiliary unit found in SBGN PD that can be hold by Entity Pool Nodes (EPNs). It informs that the entity has been duplicated: a visual indication shows that at least one other occurrence of the EPN can be found in the map. A glyph is marked by a clone marker when it is filled in it’s lower part. There are two types of clone markers: the simple clone marker (unlabeled) and the labeled clone marker.
The following example shows a simple chemical ATP with a simple clone marker.

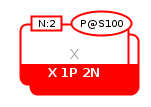
The labeled clone marker carries a label for identifying equivalent clones present elsewhere in the map. It is used for stateful EPNs to facilitate the identification when a large number of state variables are displayed making it difficult to visually identify equivalent clones. The following example represent a multimer macromolecule X that is phosphorylated (state variable P@S100) and is a dimer (unit of information N:2). The multimer X holds a labeled clone marker summarizing those information.
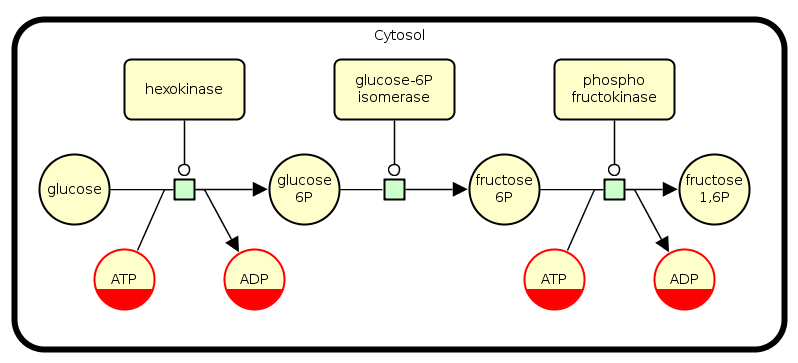
The following example shows the first three reactions that occur during the glycolysis. First, the glucose is consumed to produce glucose-6-phosphate. This reaction is catalyzed by the hexokinase enzyme and uses an ATP energy to produce ADP. The second reaction, shows the production of fructose-6-phosphate from the glucose-6-phosphate. It is catalyzed by the glucose-6-phosphate isomerase enzyme. Third, stimulated by the phosphofructokinase enzyme, the fructose-6-phosphate produces the fructose-1,6-phosphate by using an ATP that releases an ADP.
ATP and ADP are found two times in this pathway, thus both are identified by a clone marker.
Download the clone marker node image as (from the more semantic to the more generic):
[ SBGN-ML ] [ GML ] [ GraphML ] [ SVG ] [ Open Document Format ] [ PowerPoint ] [ PDF ] [ PNG ]
AF Biological Activity
by Vasundra Touré (Mar 2016)
A biological activity is an Activity Node found in SBGN AF. It is used for representing molecular activities of all types of biological entities. The glyph has a rectangle shape and carries a label representing the activity.
The following example simply shows a biological activity: Conductance activity.

A biological activity can also carry an unit of information that provides information on the nature of the entity from which the activity comes from (e.g. simple chemical, macromolecule, etc.).
The following example illustrates the binding activity (biological activity) of the macromolecule IFNG (unit of information).

It should be noted that the biological activity is not equivalent to a biological entity in itself. A biological activity can come from one biological entity, a part of an entity, or a combination of them. It is up to the users to determine how to represent it in their diagram.
The following example represents the EGF protein kinase receptor. This receptor has two activities, the binding activity that allows the extracellular part of the receptor to bind to the ligand, and the kinase activity capable of phosphorylating the downstream protein and initiating the intracellular signaling. It is possible to represent each activity in two separate nodes or to represent both activities in a single node.

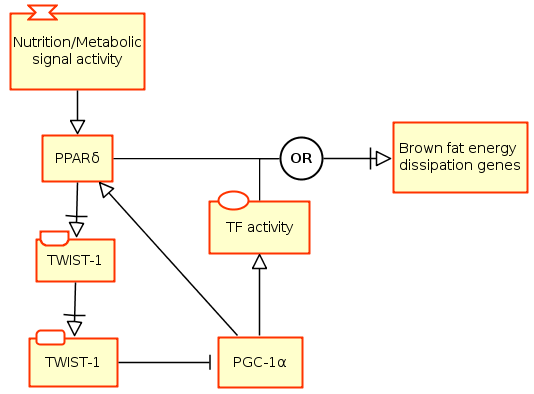
The following example is based on the Figure 7E of Pan et al [1]. It depicts the effects of nutrients and metabolic signals (shown as a perturbation) on Brown fat metabolism through PPARδ. It shows that the Twist-1, induced by the PPARδ, serves as a negative feedback regulator of PGC-1α in Brown fat metabolism. Please note the different units of information on Twist-1 activity nodes that indicate the activity from different biological materials (gene and protein).
Download the biological activity node image as (from the more semantic to the more generic):
[ SBGN-ML ] [ GML ] [ GraphML ] [ SVG ] [ Open Document Format ] [ PowerPoint ] [ PDF ] [ PNG ]
[1] D. Pan, M. Fujimoto, A. Lopes, and Y.-X. Wang, “Twist-1 Is a PPARδ-Inducible, Negative-Feedback Regulator of PGC-1α in Brown Fat Metabolism,” Cell, vol. 137, no. 1, pp. 73–86, Apr. 2009.
AF Unit Of Information for BA
by Vasundra Touré (Apr 2016)
An unit of information for a biological activity in AF illustrates the nature of the entity from which the activity comes from. It is always represented with a biological activity glyph. The unit of information can provide two types of information: the nature of the entity represented by the shape of the unit of information and the name of the entity represented by a label in the unit of information.
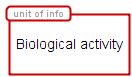
There are six types of unit of information that can characterize a biological activity, as described below:
| Name | Description | |
|---|---|---|
 |
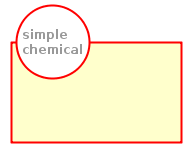
Macromolecule | Represents a very large biochemical substance, usually a protein, a nucleic acid or a complex sugar. It’s unit of information is represented by a rectangle with rounded corners. |
 |
Simple Chemical | Represents a small biochemical substance, usually an atom, a mono-atomic ion, a salt, a solid metal, etc. It’s unit of information is represented by a circular container. |
 |
Nucleic acid feature | In SBGN represents a fragment of a macromolecule carrying genetic information. It’s unit of information is represented by a rectangle whose bottom half has rounded corners. |
 |
Unspecified entity | Represents the entity type that is unknown or simply not relevant for the purposes of the maps. It’s unit of information is represented by an elliptic container. |
 |
Complex | Represents a biochemical entity composed of other biochemical entities, whether macromolecules, simple chemicals or other complexes. It’s unit of information is represented by an octagon container. |
 |
Perturbation | Represents a physical phenomena that influence a biochemical network, such as a light pulse or a change in the temperature. It can also be more complex and not well defined, for instance, glucose deprivation or stress. It’s unit of information is represented by a modified hexagon container having two opposite concave faces. |
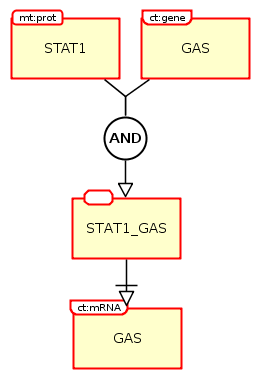
The following example shows the transcription regulation of the gene GAS by STAT1. We can see that the protein STAT1 (a macromolecule) and the gene GAS (a nucleic acid feature) needs to form a complex in order to activate the transcription of GAS mRNA.
Download the unit of information for a biological activity image as (from the more semantic to the more generic):
[ SBGN-ML ] [ GML ] [ GraphML ] [ SVG ] [ Open Document Format ] [ PowerPoint ] [ PDF ] [ PNG ]
PD Perturbing Agent
by Vasundra Touré (May 2016)
A perturbing agent is an Entity Pool Node found in PD. This glyph represents external influences carried on a biochemical network. Those influences can be the effect of well-defined physical perturbing agents, such as light pulse or a temperature change. They can also be more complex and not well-defined phenomena, for instance the outcome of a biological process or a mutation. The perturbing agent glyph is represented by a modified hexagon having two opposite concave faces. It is identified by a label and may carry a clone marker. One or more units of information can also be defined.
Units of information use a controlled vocabulary to describe the physical characteristics of a perturbing agent:
| Name | Label | SBOterm |
|---|---|---|
| Temperature | pc:T | SBO:0000147 |
| Voltage | pc:V | SBO:0000259 |
| pH | pc:pH | SBO:0000304 |
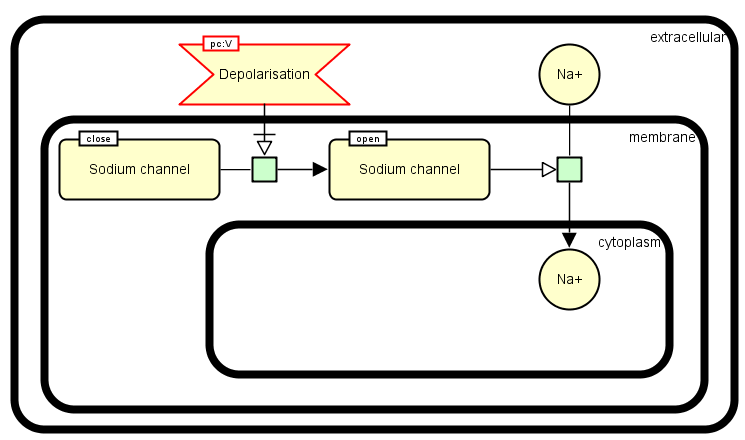
The following example shows the transport of sodium chemical Na+ into the cytoplasm of a cell via the sodium channel. The sodium channel is sensitive to the membrane potential: when the membrane is polarized, the sodium channel is closed and when the membrane is depolarized, the sodium channel is opened. In our example, the membrane is depolarized (perturbing agent). This induce the opening of sodium channel and thus the import of sodium Na+ into the cytoplasm.
Download the perturbing agent image as (from the more semantic to the more generic):
[ SBGN-ML ] [ GML ] [ GraphML ] [ SVG ] [ Open Document Format ] [ PowerPoint ] [ PDF ] [ PNG ]
PD Unspecified Entity
by Vasundra Touré (Jun 2016)
In SBGN PD, the unspecified entity is the simplest and the most generic Entity Pool Node (EPN). It defines unknown or uncertain biological elements, or simply non-relevant information for the map. An unspecified entity is represented by an elliptic shape and is identified by a label.

As it is an EPN, the unspecified entity can carry a clone marker.

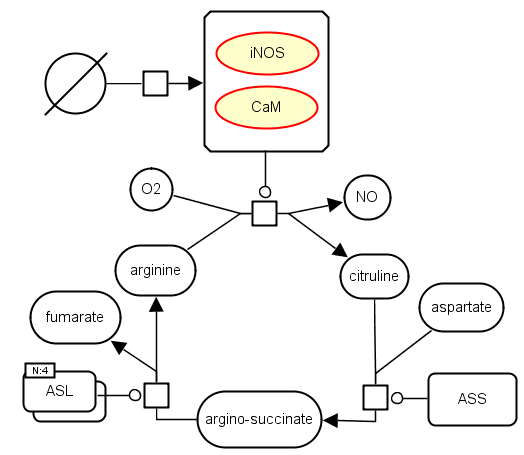
The following example shows a part of the iNOS pathway where the iNOS and CaM entities form a complex to catalyse the production of Nitric Oxide (NO) and citruline, leading to a couple of other reactions. Here, the iNOS and the CaM are represented as unspecified entities.
Download the unspecified entity glyph image as (from the more semantic to the more generic):
[ SBGN-ML ] [ GML ] [ GraphML ] [ SVG ] [ Open Document Format ] [ PowerPoint ] [ PDF ] [ PNG ]
PD Source and Sink
by Vasundra Touré (Aug 2016)
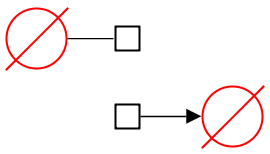
The source and sink glyphs are Entity Pool Nodes found in SBGN PD. The same symbol is used for both glyphs but the semantic differs depending on the position of the glyph upon a reaction. This symbol, inspired from the mathematical symbol “empty set”, offers to represent the creation (respectively, the disappearance) of an entity from an unspecified source (resp. sink) that we do not need or wish to explicit. A source or sink does not carry any labels neither auxiliary units.

The following figure shows on top a source glyph that is consumed and on the bottom a sink glyph that is produced. The symbol should always be linked to one edge only in a map.
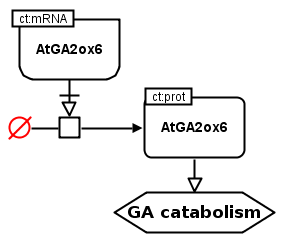
The following example is an extract of the LEC1/AFL-B3 factors and interactions with phytohormone metabolism (available in the RIMAS portal). It shows the AtGA2ox6 mRNA that is necessary to stimulate the production of the AtGA2ox6 protein, which stimulates in turn the catabolism of GA. Here the details about energy and metabolites involved in the protein’s creation is not represented. Instead, a source glyph is used stating that “a source creates” the AtGA2ox6 protein with the stimulation of the AtGA2ox6 mRNA.
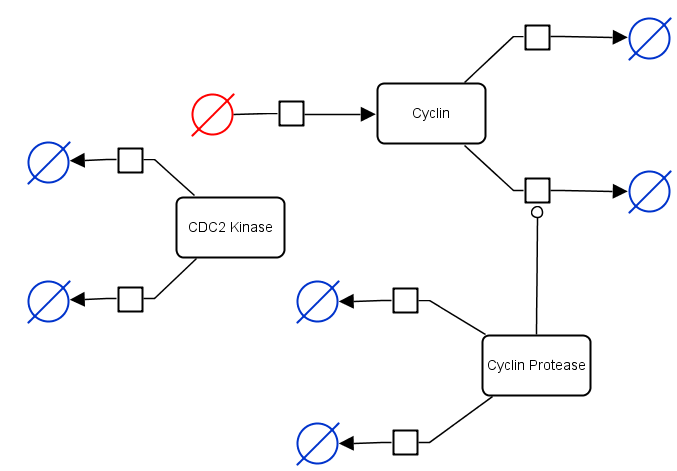
The following example shows “a minimal cascade model for the mitotic oscillator involving cyclin and cdc2 kinase” (Goldbeter et al, 1991) taken from the Biomodels database. The symbols highlighted in blue are sink glyphs and the symbol in red is a source glyph.
Download the source and sink glyph image as (from the more semantic to the more generic):
[ SBGN-ML ] [ GML ] [ GraphML ] [ SVG ] [ Open Document Format ] [ PowerPoint ] [ PDF ] [ PNG ]
PD ER and AF And
by Vasundra Touré (Sep 2016)
The glyph and is a Logical Operator that can be found in all three languages PD, ER and AF. It is used to indicate that all nodes linked as input are necessary to produce the output. The input is usually another logical operator or can be Entity Pool Nodes in PD, interactors in ER and Activity Nodes in AF. The glyph and is identified with the label “AND” and does not carry any auxiliary units.

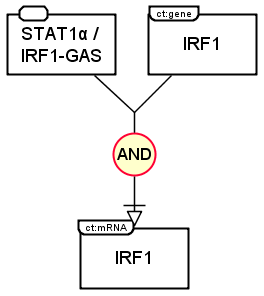
The following example shows the production of IRF1 mRNA in PD and AF languages. Both IRF1 gene and the complex formed by STAT1 protein and IRF1-GAS gene regulatory region are necessary. Thus the glyph AND links both to stimulate the process that leads to the production of IRF1 mRNA.
In Process Description language:
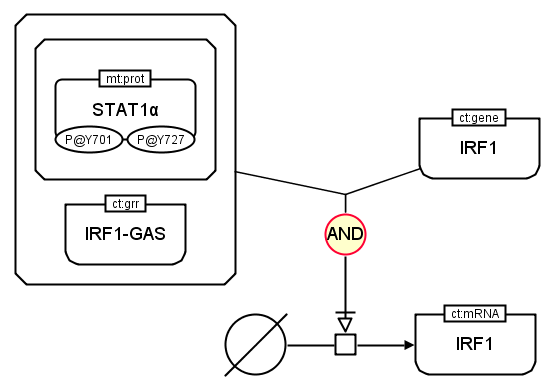
In Activity Flow language:
Download the and glyph image as (from the more semantic to the more generic):
[ SBGN-ML ] [ GML ] [ GraphML ] [ SVG ] [ Open Document Format ] [ PowerPoint ] [ PDF ] [ PNG ]
PD ER and AF Or
by Vasundra Touré (Nov 2016)
The glyph or is a Logical Operator that can be found in all the three languages PD, ER and AF. It is used to indicate that any node linked as input is sufficient to produce the output. The input is usually another logical operator or can be Entity Pool Nodes in PD, interactors in ER and Activity Nodes in AF. The glyph and is identified with the label “OR” and does not carry any auxiliary units.

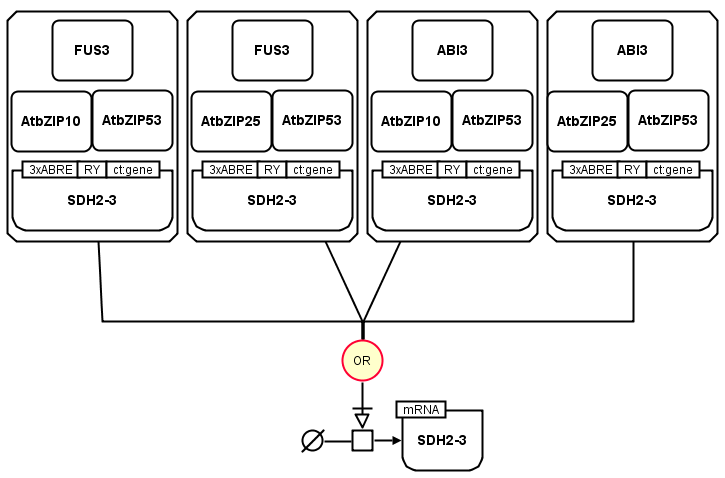
The following example shows the transcription of SDH2-3 mRNA extracted from the LEC1/AFL-B3 factors and maturation gene control (RIMAS). The transcription needs to be stimulated by at least one of the complexes showed on the map and any of these complexes is sufficient for this stimulation.
Download the or glyph image as (from the more semantic to the more generic):
[ SBGN-ML ] [ GML ] [ GraphML ] [ SVG ] [ Open Document Format ] [ PowerPoint ] [ PDF ] [ PNG ]
PD ER and AF Not
by Vasundra Touré (Dec 2016)
The glyph not is a Logical Operator that can be found in all three languages PD, ER and AF. It is used to indicate that the output only happens in the absence of the input. The input is usually another logical operator or it can be EPNs in PD, interactors in ER and ANs in AF. The glyph not is identified with the label “NOT” and does not carry any auxiliary units.

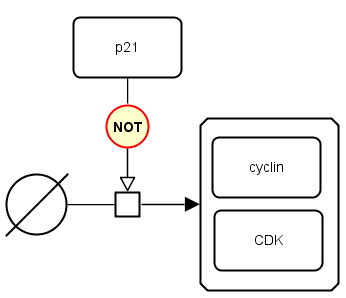
The following example shows that the production of cyclin-CDK complex is not stimulated by the protein p21.
Download the not glyph image as (from the more semantic to the more generic):
[ SBGN-ML ] [ GML ] [ GraphML ] [ SVG ] [ Open Document Format ] [ PowerPoint ] [ PDF ] [ PNG ]