WHAT IS SBGN
SPECIFICATIONS
SOFTWARE
EVENTS
FAQ
HOW TO CITE
ABOUT
CONTACT
Learning SBGN
Examples
Reference Cards
Published Maps
Symbol Highlights
Figure to SBGN
Competition
Development Wiki
Figure to SBGN
Pathways in this archive are an effort by the SBGN community to reach out to bioscience researchers. It is a redraw of a pathway diagram from a research article in SBGN format. It has to be pointed out that the SBGN redraw does not always match exactly to the original diagram. Since SBGN requires more concise and unambiguous representation of the knowledge, some modification is required based on the interpretation of the article by the SBGN curator.
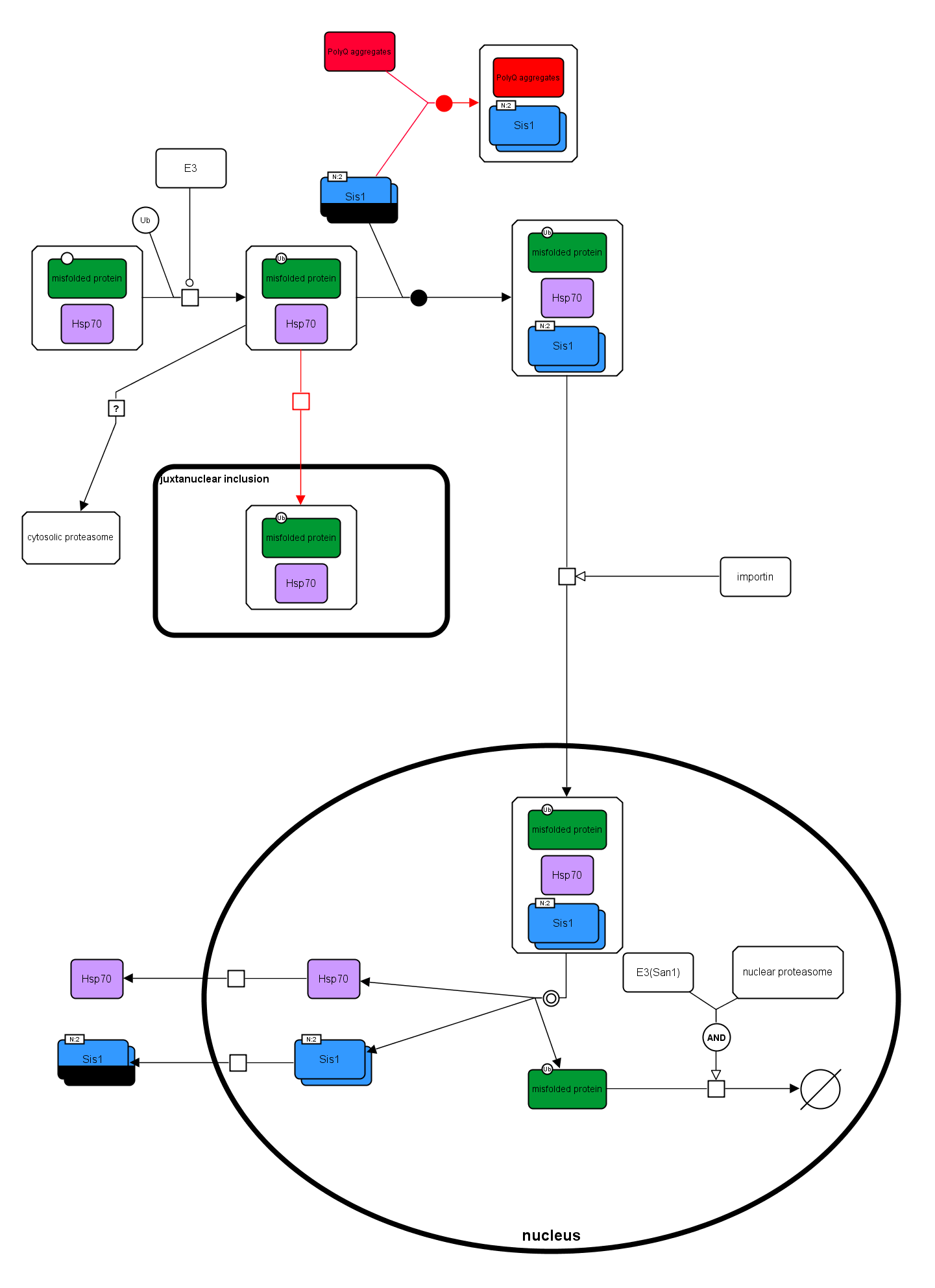
Sis1p in nuclear degradation of cytosolic misfolded proteins
An SBGN Process Desciption diagram was created based on Figure 7 of the following paper. SBGN-ED, an SBGN editing software, was used to draw the diagram.
Park S., Kukushkin Y., Gupta R., Chen T., Konagai A., Hipp M., Hayer-Hartl M., and Hartl F. (2013) PolyQ Proteins Interfere with Nuclear Degradation of Cytosolic Proteins by Sequestering the Sis1p Chaperone. Cell 154, 134-145. PMID: 23791384.
Misfolded proteins (green) are initially stabilized in a soluble state by Hsp70s (purple), and then ubiquitinated by E3 ligase (Ubr1). Recognition of misfolded protein by Sis1p dimer leads to remodeling of the initial chaperone complex, shifting from refolding to degradation mode. Sis1p stabilizes the association with Hsp70 and mediates protein import into the nucleus for proteasomal degradation. The nuclear E3 ligase San1, acting in parallel to cytosolic E3 ligases, contributes to efficient ubiquitylation. When the cell contains PolyQ aggregate, it will deplete Sis1 by binding it, and thus preventing the complex from entering the degradation mode. See the article for more details.
There are some modifications in the SBGN diagram compared to the original one.
-
In the original diagram, there is an inhibition arrow from the PolyQ aggregate. This is actually ambiguous. The PolyQ aggregate does not have the inhibition activity. Rather, it is the process of depleting of Sis1 by PolyQ aggregate that inhibits the binding of Sis1 to the misfolded protein. Therefore, this inhibition arrow is omitted in the SBGN diagram.
-
A logic gate is used to show that both nuclear E3 ligase and nuclear proteamsome are required to degrade the misfolded protein.
Designed by:
Huaiyu Mi, PhD, Associate Professor of Research, University of Southern California, Keck School of Medicine.
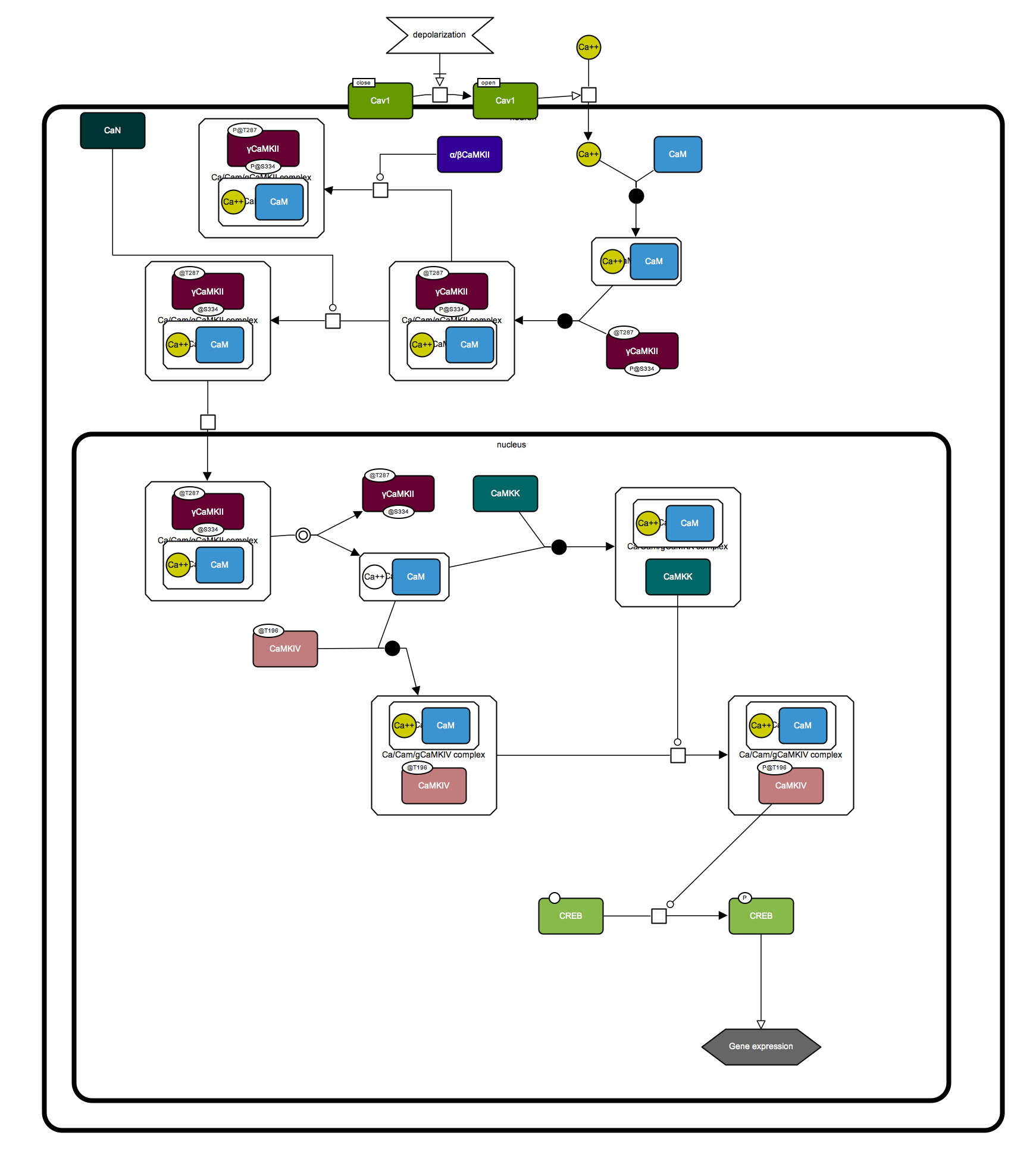
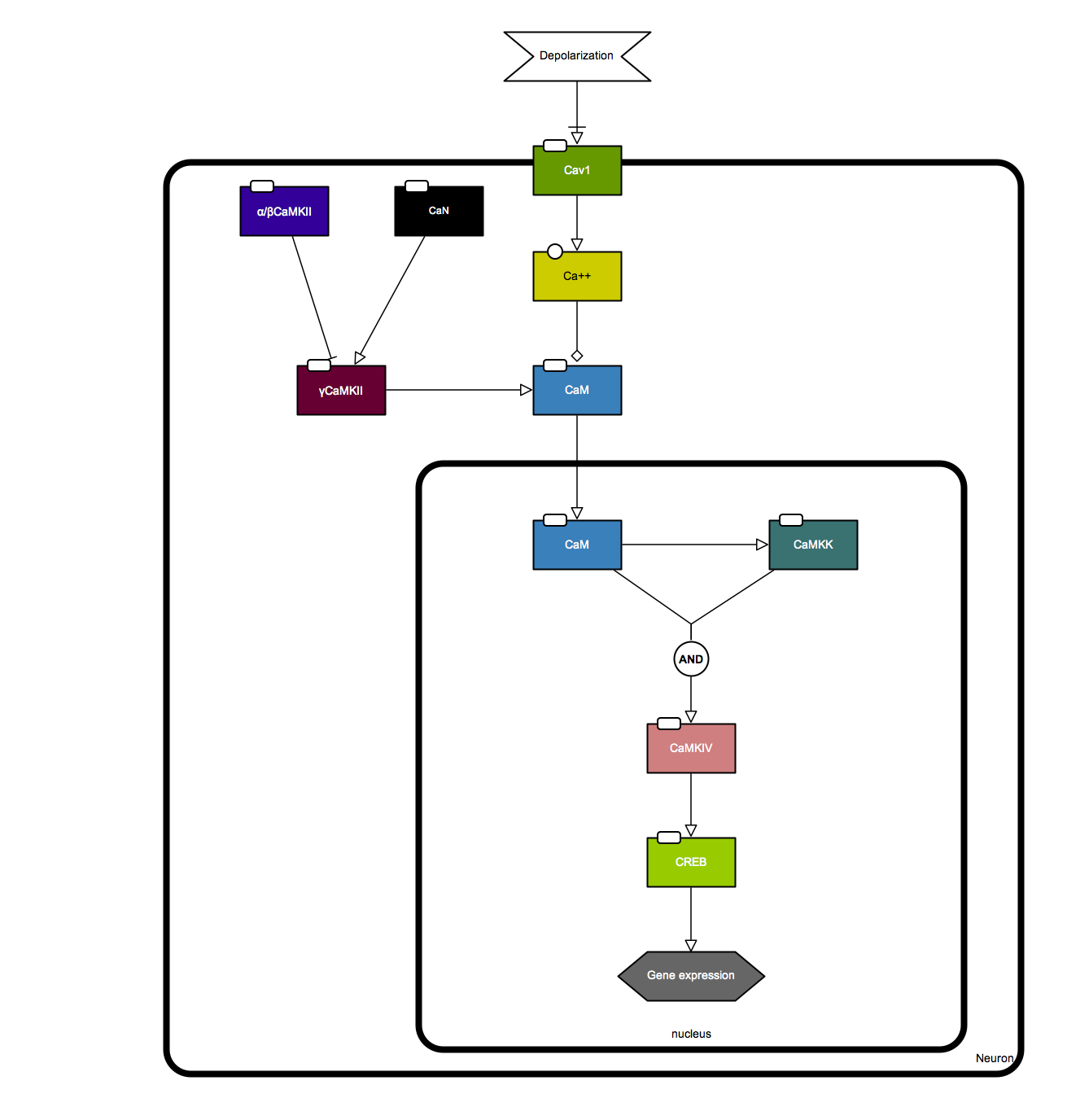
γCaMKII shuttles Ca²⁺/CaM to the nucleus to trigger CREB phosphorylation and gene expression
An SBGN Process Desciption and Activity Flow diagrams were created based on the Graphical Abstract of the following paper. SBGN-ED, an SBGN editing software, was used to draw the diagram.
Ma H, Groth RD, Cohen SM, Emery JF, Li B, Hoedt E, Zhang G, Neubert TA, Tsien RW. (2014) γCaMKII shuttles Ca²⁺/CaM to the nucleus to trigger CREB phosphorylation and gene expression. Cell 159,281-294. PMID: 25303525.
Activity-dependent CREB phosphorylation and gene expression are critical for long-term neuronal plasticity. Local signaling at CaV1 channels triggers these events, but how information is relayed onward to the nucleus remains unclear. Here, we report a mechanism that mediates long-distance communication within cells: a shuttle that transports Ca(2+)/calmodulin from the surface membrane to the nucleus. We show that the shuttle protein is γCaMKII, its phosphorylation at Thr287 by βCaMKII protects the Ca(2+)/CaM signal, and CaN triggers its nuclear translocation. Both βCaMKII and CaN act in close proximity to CaV1 channels, supporting their dominance, whereas γCaMKII operates as a carrier, not as a kinase. Upon arrival within the nucleus, Ca(2+)/CaM activates CaMKK and its substrate CaMKIV, the CREB kinase. This mechanism resolves long-standing puzzles about CaM/CaMK-dependent signaling to the nucleus. The significance of the mechanism is emphasized by dysregulation of CaV1, γCaMKII, βCaMKII, and CaN in multiple neuropsychiatric disorders.
Below is the SBGN PD map of the pathway.
Designed by:
Huaiyu Mi, PhD, Associate Professor of Research, University of Southern California, Keck School of Medicine.
Here is the SBGN AF map of the pathway.
Designed by:
Huaiyu Mi, PhD, Associate Professor of Research, University of Southern California, Keck School of Medicine.