
WHAT IS SBGN
SPECIFICATIONS
SOFTWARE
EVENTS
FAQ
HOW TO CITE
ABOUT
CONTACT
Learning SBGN
Examples
Reference Cards
Published Maps
Symbol Highlights
Figure to SBGN
Competition
Development Wiki
Welcome to the global portal for documentation, news, and other information about the Systems Biology Graphical Notation (SBGN) project, an effort to standardise the graphical notation used in maps of biological processes.
Cool visualisation
|
PD map of the Drosophila cell cycle, doi:10.1371/journal.pcbi.1005740 
|
AF map of protein precursor processing, doi:10.1002/psp4.12155 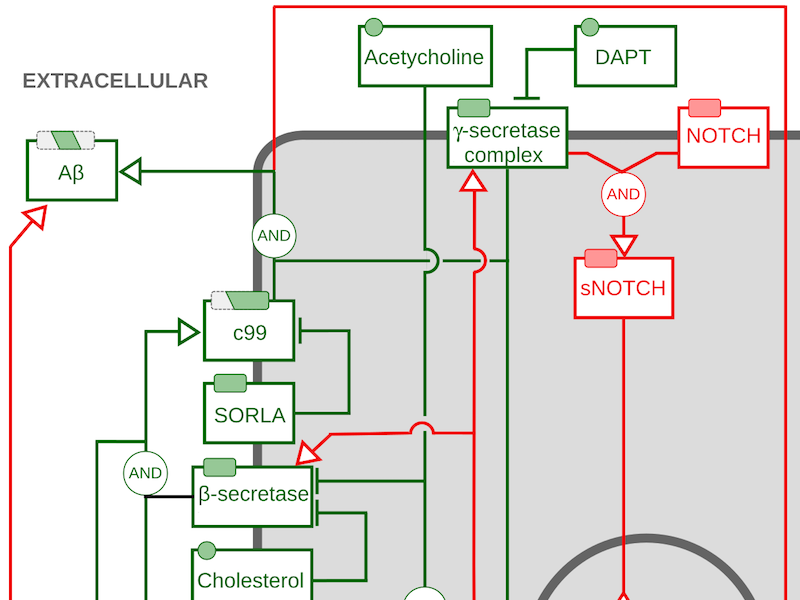
|
ER map of CaMKII regulation by calmodulin, doi:10.1371/journal.pone.0029406 
|
|---|
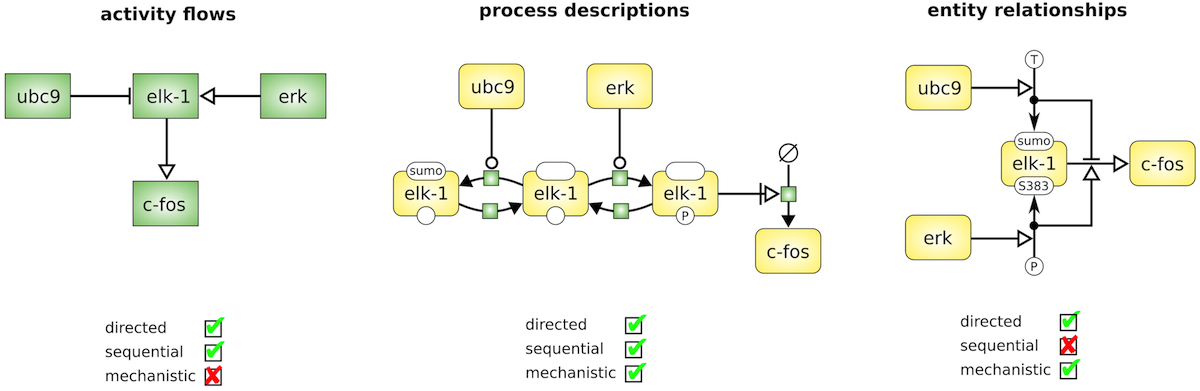
Exact meaning of all symbols
Below, the same biological system (Le Novère, 2015, doi:10.1038/nrg3885) is shown in different languages of SBGN: Activity Flow (AF), Process Description (PD) and Entity Relationship (ER).
Exchange and storage format SBGN-ML and software library
Various tools can export an import SBGN visualization in the XML-based file format SBGN-ML. SBGN-ML can be processed and generated by the standard library libSBGN One can compare the rendering of SBGN-ML by different software packages on as previously done libSBGN Render Comparison.
Tools and Databases
Databases visualizing pathways as SBGN: Reactome*, PANTHER Pathway*, BioModels database, Pathway Commons*, Atlas of Cancer Signalling Networks*, Path2Models*, and more (*exporting SBGN-ML).
Modeling software that draw PD diagrams: CellDesigner, BioUML.
Editors that draw SBGN diagrams and export/import SBGN-ML: KrayonForSbgn (PD), Newt Editor (PD, AF), Vanted/SBGN-ED (PD, AF, ER), PathVisio (PD, AF, ER), yEd/ & ySBGN (PD, AF), and more.
Tools that convert different formats to SBGN: ChiBE (BioPAX → SBGN), KEGGtranslator (KEGG-ML → SBGN), CySBGN (SBML → SBGN), SBGNTikZ (LaTex → SBGN), and more.
Tools that visualize PD diagrams: BIOCHAM, COPASI, JWS Online and more.
SBGN is the work of many people. It would not have been possible without the generous support of multiple organizations over the years, for which we are very thankful.